DREAM challenges¶
Contents
DREAM2¶
D2C1¶
D2C2 scoring function
Class imlemented in Python based on original code in MATLAB from Gustavo A. Stolovitzky.
D2C2¶
D2C2 scoring function.
Implementation in Python based on a MATLAB code from Gustavo A. Stolovitzky
-
class
D2C2(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D2C2 challenge
from dreamtools import D2C2 s = D2C2() filename = s.download_template() s.score(filename)
constructor
-
score(filename)[source]¶ Returns statistics (e.g. AUROC)
Parameters: filename (str) – a valid filename as returned by download_template()
-
D2C3¶
D2C3 scoring functions
The original algorithm was developed in MATLAB by Gustavo Stolovitzky
-
class
D2C3(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D2C3 challenge
from dreamtools import D2C3 s = D2C3() subname = "DIRECTED-UNSIGNED_qPCR" filename = s.download_template(subname) s.score(filename, subname)
There are 12 gold standards and templates. There are scored independently (6 for the chip case and 6 for the qPCR).
Although there is no sub-challenge per se, there are 12 different templates so we use the template names as sub-challenge names
constructor
-
download_goldstandard(subname=None)[source]¶ Returns one of the 12 gold standard files
Parameters: subname – one of the sub challenge name. See sub_challenges
-
sub_challenges= None¶ sub challenges (12 different values)
-
D2C4¶
D2C4 scoring function
original code in MATLAB by Gustavo Stolovitzky
-
class
D2C4(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D2C4 challenge
from dreamtools import D2C4 s = D2C4() subname = 'DIRECTED-UNSIGNED_InSilico1' filename = s.download_template(subname) s.score(filename, subname)
constructor
-
sub_challenges= None¶ 12 different sub challenges
-
D2C5¶
D2C5 scoring functions
Original code in MATLAB by Gustavo Stolovitzky
DREAM3¶
D3C1¶
D3C1 scoring function
Original matlab code from Gustavo A. Stolovitzky and Robert Prill.
D3C2¶
D3C2 scoring function
Implemented after an original MATLAB code from Gustavo Stolovitzky and Robert Prill.
-
class
D3C2(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D3C2 challenge
from dreamtools import D3C2 s = D3C2() filename = s.download_template('cytokine') s.score(filename, 'cytokine') filename = s.download_template('phospho') s.score(filename, 'phospho')
Data and templates are downloaded from Synapse. You must have a login.
constructor
D3C3¶
D3C3 scoring function
Original matlab version (Gustavo A. Stolovitzky, Ph.D. Robert Prill) translated into Python by Thomas Cokelaer.
-
class
D3C3(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D3C3 challenge
from dreamtools import D3C3 s = D3C3() filename = s.download_template() s.score(filename)
Data and templates are downloaded from Synapse. You must have a login.
Note
the spearman pvalues are computed using R and are slightly different from the official code that used matlab. The reason being that the 2 implementations are different. Pleasee see cor.test in R and corr() function in matlab for details. The scipy.stats.stats.spearman has a very different implementation for small size cases.
constructor
D3C4¶
Implementation in Python from Thomas Cokelaer. Original code in matlab (Gustavo Stolovitzky and Robert Prill).
-
class
D3C4(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D3C4 challenge
from dreamtools import D3C4 s = D3C4() filename = s.download_template(10) s.score(filename)
Note
AUROC/AUPR and p-values are returned for a given sub-challenge. In the DREAM LB, the 5 networks are combined together. We should have same implemntatin as in D4C2
constructor
DREAM4¶
D4C1¶
D4C1 scoring function
Based on an original matlab code from Gustavo A. Stolovitzky, and Robert Prill.
D4C2¶
D4C2 scoring function
From an original code in matlab (Gustavo Stolovitzky and Robert Prill).
D4C3¶
D4C3 scoring function
Based on Matlab script available on https://www.synapse.org/#!Synapse:syn2825304, which is an original code from Gustavo A. Stolovitzky and Robert Prill.
-
class
D4C3(verbose=True, download=True, edge_count=None, cost_per_link=0.0827, **kargs)[source]¶ A class dedicated to D4C3 challenge
from dreamtools import D4C3 s = D4C3() filename = s.download_template() s.edge_count = 20 s.score(filename)
Data and templates are inside Dreamtools.
Note
A parameter called cost_per_link is hardcoded for the challenge. It was compute as min {Prediction Score / Edge Count} amongst all submissions. For this scoring function,
cost_per_linkis set to 0.0827 and may be changed by the user.constructor
Parameters:
DREAM5¶
D5C1¶
D5C1 scoring function
From an original matlab code from Gustavo A. Stolovitzky, Robert Prill
D5C2¶
D5C2 challenge scoring functions
Based on TF_web.pl (perl version) provided by Raquel Norel (Columbia University/IBM) also used by the web server http://www.ebi.ac.uk/saezrodriguez-srv/d5c2/cgi-bin/TF_web.pl
This implementation is independent of the web server.
-
class
D5C2(verbose=True, download=True, tmpdir=None, Ntf=66, **kargs)[source]¶ A class dedicated to D5C2 challenge
from dreamtools import D5C2 s = D5C2() # You can get a template from www.synapse.org page (you need # to register) filename = s.download_template() s.score(filename) # takes about 5 minutes s.get_table() s.plot()
Data and templates are downloaded from Synapse. You must have a login.
constructor
Parameters: - Ntf – not to be used. Used for fast testing and debugging
- tmpdir – a local temporary file if provided.
-
compute_statistics()[source]¶ Returns final results of the user predcition
Returns: a dataframe with various metrics for each transcription factor. Must call
score()before.
-
download_template()[source]¶ Download a template from synapse into ~/config/dreamtools/dream5/D5C2
Returns: filename and its full path
-
get_table()[source]¶ Return table with user results from the user and participants
There are 14 participants as in the Leaderboard found here https://www.synapse.org/#!Synapse:syn2887863/wiki/72188
Returns: a dataframe with different metrics showing performance of the submission with respect to other participants. table = s.get_table() with open('test.html', 'w') as fh: fh.write(table.to_html(index=False))
D5C3¶
D5C3 scoring function
Original matlab code from Gustavo A. Stolovitzky, Robert Prill, Ph.D. sub challenge B original code in R from A. de la Fuente
-
class
D5C3(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D5C3 challenge
from dreamtools import D5C3 s = D5C3() filename = s.download_template() s.score(filename)
Data and templates are downloaded from Synapse. You must have a login.
3 subchallenges (A100, A300, A999) but also 3 others simpler with B1, B2, B3
For A series, 5 networks are required. For B, 3 are needed.
constructor
D5C4¶
D5C4 scoring function
Based on original matlab code from Gustavo A. Stolovitzky and Robert Prill
DREAM6¶
D6C1¶
D6C1 scoring function
scoring author: bobby prill
D6C2¶
D6C2 scoring function
See D7C1
D6C3¶
D6C3 scoring function
Based on Pablo’s Meyer Matlab code.
-
class
D6C3(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D6C3 challenge
from dreamtools import D6C3 s = D6C3() filename = s.download_template() s.score(filename)
Absolute score in the Pearson coeff but other scores such as chi-square and rank are based on the 21st participants.
Pearson and spearman gives same values as in final LB but X2 and R2 are slightly different. Same results as in the original matlab scripts so the different with the LB is probably coming fron a different set of predictions files, which is stored in ./data/predictions and was found in http://genome.cshlp.org/content/23/11/1928/suppl/DC1
The final score in the official leaderboard computed the p-values for each score (chi-square, r-square, spearman and pearson correlation coefficient) and took -0.25 log ( product of p-values) as the final score.
constructor
D6C4¶
Original scoring function: Kelly Norel
DREAM7¶
D7C1¶
DREAM7 Challenge 1 (Parameter estimation and network topology prediction)
| References: | |
|---|---|
| Publications: |
-
class
D7C1(verbose=True, download=True, path='submissions', **kargs)[source]¶ DREAM 7 - Network Topology and Parameter Inference Challenge
Here is a quick example on calling the scoring methods:
from dreamtools import D7C1 s = D7C1() s.score_model1_timecourse(filename) s.score_model1_parameters(filename) s.score_topology(filename)
This class provides 3 main scoring functions:
Each takes as an input a valid submission as described in the official synapse page.
Templates are also provided within the source code on github dreamtools in the directory dreamtools/dream7/D7C1/templates.
D7C1 scoring function are also included in the standalone code dreamtools-scoring.
For the details of the scoring functions, please refer to the paper (see module documentation) Some details are provided in the methods themselves as well.
There are other methods (starting with leaderboard) that should not be used. Those are draft version used to compute pvalues and report scores as in the final leaderboard.
Note
the scoring functions were implemented following Pablo Meyer’s matlab codescore_dream7_c1s1.m
For admin only: put the submissions in ./submissions/ directory and call the :meth:
constructor
Parameters: path – path to a directory containing submissions Returns: -
download_template(name)[source]¶ Return filename of a template
Parameters: name (str) – one in ‘topology’, ‘parameter’, ‘timecourse’
-
leaderboard()[source]¶ Computes all scores for all submissions and returns dataframe
Returns: dataframe with scores for each submissions for the model1 (parameter and timecourse) and model2 (topology)
-
leaderboard_compute_score_parameters_model1()[source]¶ Computes all scores (parameters model1)
Returns: Nothing but fills scores.For the metric, see
score_model1_parameters().See also
-
leaderboard_compute_score_timecourse_model1(startindex=10, endindex=39)[source]¶ Computes all scores (timecourse model1)
Returns: Nothing but fills scoresFor the metric, see
score_model1_parameters().Note that endindex is set to 39 so it does not take into account last value at time=20 This is to be in agreement with the implemenation used in the final leaderboard
https://www.synapse.org/#!Synapse:syn2821735/wiki/71062
If you want to take into account final point, set endindex to 40
-
leaderboard_compute_score_topology()[source]¶ Computes all scores (topology) for loaded submissions
For the metric, see
score_topology().Returns: fills scores.See also
-
load_submissions()[source]¶ Load a bunch of submissions to be found in the submissions directory
The directory name is defined in
pathReturns: nothing. Populates dataattribute andteam_names.
-
score(filename, subname=None)[source]¶ Return score for a given sub challenge
Parameters: filename (str) – input filemame. Returns: name of a sub_challenge. See sub_challengesattribute.
-
score_model1_parameters(filename)[source]¶ Return distance between submission and gold standard for parameters challenge (model1)
Parameters: filename – must be valid templates Returns: score (distance) >>> from dreamtools import D7C1 >>> s = D7C1() >>> filename = s.download_template('parameter') >>> s.score(filename, 'parameter') 0.022867555017785129
The score is computed using the square of the ratio of the user prediction and the gold standard. Taking the mean of the log10 :
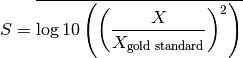
-
score_model1_timecourse(filename)[source]¶ Returns distance between prediction and gold standard (model1)
Parameters: filename – must be valid templates Returns: score (distance) >>> from dreamtools import D7C1 >>> s = D7C1() >>> filename = s.download_template('timecourse') >>> s.score_model1_timecourse(filename) 0.0024383612676804048
There are 3 time courses to be predicted. The score for each time course is
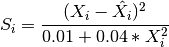
where
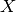 is the gold standard and
is the gold standard and 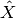 the prediction.
and final score is just the average across the 3 time courses.
the prediction.
and final score is just the average across the 3 time courses.
-
score_topology(filename)[source]¶ Standalone version of the network topology scoring
Parameters: filename (str) – >>> from dreamtools import D7C1 >>> s = D7C1() >>> filename = s.download_template('topology') >>> s.score(filename, 'topology') 12
Scoring details: The challenge requests predictions for 3 missing links, knowing that a gene can regulate up to two genes when they are in the same operon, 6 gene interactions have to be indicated by the participants (3 links*2 genes) and whether these interactions are activating (+) or repressing (-).
For each of the predicted links i=1,2,3, we define a score:
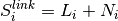
where
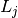 is 6 if the nature of
the regulation iscorrect (that is, the source gene, the sign of the connection, and
the destination gene are all correct) and
is 6 if the nature of
the regulation iscorrect (that is, the source gene, the sign of the connection, and
the destination gene are all correct) and 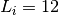 if the link
regulates an operon composed of two genes and both connections are
correct. If
if the link
regulates an operon composed of two genes and both connections are
correct. If 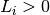 then
then 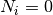 .
.In case a link is NOT correctly predicted (
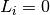 )
) 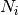 is defined as follows. It is increased by 1 for each correctly
regulated gene, 2 if the regulated gene and the nature of the
regulation (i.e +/-) are correct and 1 if the regulator gene is
correct
is defined as follows. It is increased by 1 for each correctly
regulated gene, 2 if the regulated gene and the nature of the
regulation (i.e +/-) are correct and 1 if the regulator gene is
correctThe gold standard contains 3 lines similar to
5 + 7 + 11
It means gene 5 positively regulates gene 7 and gene 11. If a prediction is
5 + 7 + 2
Then L =6. If the prediction is
2 + 7 + 2
L = 0 so N may be updated. Here the regulon (2) is not correct, However, one gene (7) is correctly predicted with the good sign so N = 2.
-
D7C2¶
-
class
D7C2(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D7C2 challenge
from dreamtools import D7C2 s = D7C2() filename = s.download_template() s.score(filename)
Data and templates are downloaded from Synapse. You must have a login.
as R objects implementing a function called customPredict() that returns a vector of risk predictors when given a set of feature data as input. The customPredict()
constructor
D7C3¶
D7C4¶
Original code for challenge B translted from Mukesh Bansal Sub challenge A is currently a wrapping of a perl code provided by Jim Costello
-
class
D7C4(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D7C4 challenge
from dreamtools import D7C4 s = D7C4() filename = s.download_template() s.score(filename)
Data and templates are downloaded from Synapse. You must have a login.
# columns represent the probabilistic c-index of the given team for each drug. # following the columns of teams are 5 columns which are used for calculating the overall team score # |-> Test_data = the probabilistic c-index for the experimentally determined test data scored against itself # |-> Mean Null Distribution = a set of 10,000 random predictions were scored to create the null distribution, of which this column represents the mean # |-> SD Null Distribution = a set of 10,000 random predictions were scored to create the null distribution, of which this column represents the standard deviation # |-> z-score of test data to null = score of the test data minus the mean of the null distribution divided by the standard deviation of the null distribution # |-> weight of drug (normalized z-score) = the z-score normalized by the largest z-score across all 31 drugs. # to calculate your team overall score, simply mulitple the score of all drugs by the corresponding weight. Divide the sum of these weighted scores by the sum of the weights
constructor
This challenge uses PERL script that requires specific packages.
First, you need cpanm tools (http://search.cpan.org/dist/App-cpanminus/)
Under Fedora 23:
sudo dnf install perl-App-cpanminusThen, install the dependencies that will be required
sudo cpanm install Math::Libm sudo cpanm install Algorithm::Pair::Best2 sudo cpanm install Digest::SHA1 sudo cpanm install Tk sudo cpanm install Games::Go::AGATournfinally install the Games-go-GoPair.tar.gz package stored in dreamtools github repositotry in dreamtools/dreamt7/D7C4/misc:
cd dreamtools/dream7/D7C4/misc tar xvfz Games-Go-GoPair-1.001.tar.gz cd Games-Go-GoPair-1.001 perl Makefile.PL make sudo make install
DREAM8¶
D8C1¶
This module provides utilities to compute scores related to HPN-Dream8
It can be used and should be used indepently of Synapse altough for testing, data sets may be downloading from synapse if you don’t have any local files to play with.
Here is an example related to the Network subchallenge:
>>> from dreamtools.dream8.D8C1 import scoring
>>> s = scoring.HPNScoringNetwork()
>>> s.compute_all_descendant_matrices()
>>> s.compute_all_rocs()
>>> s.compute_all_aucs()
https://www.synapse.org/#!Synapse:syn1720047/wiki/60530
-
class
HPNScoringNetwork(filename=None, verbose=False, true_descendants=None)[source]¶ Class to compute the score of a Network submission
A user will provide a ZIP file that contains 65 files: 32 EDA, The 32 files should be tagged with the 32 combos of cell lines and ligands. To create an instance of HPNScoringNetwork, type:
s = HPNScoringNetwork("TeamName-Network.zip") # or later s = HPNScoringNetwork() s.load_submission("TeamName-Network.zip") s.get_auc_final_scoring() # as in the challenge ignoring some regimes
You then need to specifically load the EDA files. This may be done with
load_all_eda_files_from_zip():s.load_all_eda_files_from_zip()
The content of the ZIP file can be validated using the
validation()method.:s.validation()
Each EDA and SIF file must be a complete graph where all species correspond to the CSV files provided on the synapse web page. The size of the network varies depending on the cell line.
Each EDA file that contains score on each edge and first needs to be transformed into a descendancy matrix. This is achieve via
compute_descendant_matrix()and/orcompute_descendant_matrix()methods.:s.compute_all_descendant_matrices()
From each matrix, we’d like to compare a specific row (corresponding to mTOR) to the true scores that are expected. The true descendant for each combinaison of cell line and ligand are provided and loaded in the constructor via
load_true_descendants_from_zip(), which can be called at any time.Parameters: filename (str) – -
compute_all_aucs()[source]¶ Computes all AUC
This function can be called once EDA files are loaded and all descendant matrices have been computed as well.
In theory, one should compute ROC and then AUC but this function recomputes ROC since it is fast to compute.
-
compute_all_descendant_matrices()[source]¶ Compute all descendancy matrices
For each cell line and ligand, the matrix is stored in the
edge_scoresdictionary.See also
-
compute_all_rocs()[source]¶ Computes all ROC curves
This function can be called once EDA files are loaded and all descendant matrices have been computed as well.
-
compute_auc(roc)[source]¶ Compute AUC given a ROC data set
Parameters: roc (str) – The roc data structure must be a dictionary with “tpr” key. Could be an variable returned by compute_roc().
-
compute_descendant_matrix(cellLine, ligand)[source]¶ Computes the descendancy matrix for a given cell line and ligand
Parameters: Note
we use a cython module to conmpute the matrix. This function is the bottle neck of the entire procedure to compute the score. This is especially important to estimate te null distribution of the AUCs. Using Cython does not improve much the performance (80%) but it improves it...
See also
-
compute_roc(cellLine, ligand)[source]¶ Compute the ROC curve
Parameters: - scores – list of scores (probabilities)
- classes – list of classes (true values)
See also
-
compute_score(validation=True)[source]¶ Computes the final score that is the average over the 32 AUCs
This function compute the final score. First, il loads all EDA files provided by the participany (from the ZIP file). Then, it computes the 32 descendant matrices. Finally, it computes the 32 ROCS and AUCS. The scores is for now based on the z-score. Since scores must be between 0 and 1, where 0 is the best, we will need to normalise.
Parameters: validation (bool) – perform validation of the input ZIP file
-
get_auc_final_scoring()[source]¶ This function returns the mean AUC using only official ligands as used in final scoring and collaborative rounds.
The individual AUCs must be computed first with
compute_all_aucs()or .:>>> s = scoring.HPNScoringNetwork(filename) >>> auc = s.get_auc_final_scoring()
-
get_mean_and_sigma_null_parameters()[source]¶ Retrieve mean and sigma for 32 combi from a null AUC distribution
-
get_null_distribution(sample=100, cellLine='BT20', ligand='EGF', store_rocs=False, distr='uniform')[source]¶ Computes the null distribution for a given combinaison
- Creates a uniformly distribution of a EDA file and stores it in the edge_score attribute.
- recompute the corresponding descendancy matrix
- Get the corresponding true prediction
- compute the ROC and AUC
Parameters: Returns: rocs and aucs (rocs is set to [] for debugging)
from dreamtools.dream8.D8C1 import scoring from pylab import clf, plot, hist, grid, pi, exp, sqrt, mean, std s = scoring.HPNScoringNetwork() rocs, aucs, auprs = s.get_null_distribution(100) mu = mean(aucs) sigma = std(aucs) clf() res = hist(aucs,50, normed=True) plot(res[1], 1/(sigma * sqrt(2 * pi)) * exp( - (res[1] - mu)**2 / (2 * sigma**2) ), linewidth=2, color='r') grid()
-
load_eda_file(filename, local=False)[source]¶ Loads scores from one EDA file
Parameters: filename – here filename should be one of the filename to be found within the ZIP file! This is not a standard file system (See note). Input data is EDA format that is:
A 1 B = 0.4 A 1 C = 0.5
It containts edges such that the final graph is complete and a matrix can be built with column1 as the rows and column2 as the columns. The values being tmade from the fifth column. Second and fourth are ignored.
loaded data is stored in
dataas a numpy matrix.Note
to overwrite the input ZIP file, use
loadZIPFile()
-
plot_all_rocs(cellLines=None)[source]¶ Plots all 32 ROC once scores/rocs have been computed
from pylab import clf, plot, hist, grid from dreamtools.dream8.D8C1 import scoring import os s = scoring.HPNScoringNetwork() from dreamtools import D8C1 filename = D8C1().download_template('SC1A') s.load_submission(filename) s.compute_score() s.plot_all_rocs()
-
test_synapse_id= 'syn1971273'¶
-
true_synapse_id= 'syn1971278'¶
-
-
class
HPNScoring(verbose=True)[source]¶ Base class common to all scoring classes
The HPN challenges use data from 32 types of combinaison of cell lines (4) and ligands (8). This class provides aliases to:
- valid cell lines (
valid_cellLines) - valid ligands (
valid_ligands) - expected length of the vectors for each cell line
(
valid_length) - indices of the row vectors containing the mTOR species within the
descendancy matrices
(
mTor_index)
Note
all matrices and vectors are sorted according to a hard-coded list of species for a combinaison of cell line and ligand. The species are indeed sorted alphabetically following hhe same order as in the original CSV files containing the data sets.
In addition, it the
scoreattributes can be used to store the score computed bycompute_score().All classes that need to compute scores require a data file submitted by a participant. We enforce the usage of ZIP file, which can be loaded by using
loadZIPFile().-
error(message)[source]¶ If you want to raise an error, use this method.
It raises a ScoringException and set the
exceptionattribute. The message is stored in exception.value If called, the :attr:` ` is set to “INVALID” and the score is set to 1 (worst score).
-
load_species()[source]¶ Loads names of the expected phospho names for each cell line from the synapse files provided to the users
-
mTor_index= None¶ indices of the mTOR species in the different cell lines within
-
score¶ R/W attribute to store the score (in [0,1] only)
-
valid_cellLines= None¶ List of valid cell lines (e.g, BT20)
-
valid_length= None¶ length of the vectors to be found within each cell line
-
valid_ligands= None¶ List of valid ligands (e.g, EGF)
- valid cell lines (
-
class
HPNScoringNetworkInsilico(filename=None, verbose=False)[source]¶ Scoring class for HPN DREAM8 Network Insilico sub challenge
This class retrieves the true graph and a test example from synapse.
from dreamtools.dream8.D8C1 import HPNScoringNetworkInsilico s = HPNScoringNetworkInsilico() import os filename = s.download_template("SC1B") s.read_file(filename)
Note
If you want to test your own local file, provide a filename.
-
auc¶
-
get_null_auc_aupr(N)[source]¶ Get null distribution of the AUCs and AUPRs
Parameters: N (int) – number of samples Returns: tuple made of 2 lists: the AUCc ad AUPRs
-
get_zscore()[source]¶ Returns scores for the current submission
Returns: a single value based on the assumption that the distribution of the NULL AUC follows a gaussian distribution with parameters that are hardcoded as mu=0.497404 and std=0.037436. aucs2, auprs2 = s.get_null_auc_aupr(500000) scipy.stats.gamma.fit([x for x in auprs if numpy.isnan(x)==False]) scipy.stats.norm.fit(aucs)
-
plot_null_distribution(aucs=None, auprs=None, N=10000)[source]¶ Plots the null distribution of the AUCs
from dreamtools.dream8.D8C1 import HPNScoringNetworkInsilico from dreamtools import D8C1 import os s = HPNScoringNetworkInsilico() filename = D8C1().download_template('SC1B') s.read_file(filename) aucs, auprs = s.get_null_auc_aupr(1000) s.plot_null_distribution(aucs) from pylab import xlim xlim([0.35,0.65])
-
test_synapse_id= 'syn1973430'¶
-
true_synapse_id= 'syn1976597'¶
-
-
class
HPNScoringNetwork_ranking[source]¶ This class is used to compute the ranks of the different participants based on an average rank over the 32 combinaisons of cell line and ligands.
s = HPNScoringNetwork(filename="file1zip") s.compute_all_aucs() sall = HPNScoringNetwork_all() # s.aucs is a list where each element is a dictionary of sall.add_auc(s1.auc, "team1") # let us build aucs2 = copy.deepcopy(s.auc) for c in s.valid_cellLines: for l in s.valid_ligands: auc2[c][l] = numpy.random.uniform(0.5,0.7) sall.add_auc(s2.auc, "team2") sall.get_ranking() {'team1': 1.96875, 'team2': 1.03125}
This class is independent of HPNSCoringNetwork. However, it takes as input the returned values of HPNScoringNetwork.compute_all_auc()
-
class
HPNScoringPrediction(filename=None, version=2, verbose=False)[source]¶ -
compute_all_rmse()[source]¶ Some species were removed on purpose during the analysis
Those are hardcoded. To compute null distribution, we can keep all the species, in which case, _version parameter must be 0 must be set to False.
-
create_random_data()[source]¶ Here, we don’t want the true prediction that contains only what is requested (AZD8055) but the orignal training data with 2 or 3 inhibitors such as GSK and PD17 so that we can shuffle them.
We want to select for a given cell line and phosphos a data set to fill at a given time. The datum is selected accross the 8 stimuli, inhibitors +DMSO, and time points.
TAZ and FOX were asked to be excluded so this cause some trouble now but some user preidction still include them. Should add a if statement to ignore them. Does not matter to compute the null distribution
-
get_null(N=100, tag='sc2a')[source]¶ s = HPNScoringPrediction() nulls = s.get_null(1000) # the nulls contains the 4 cell lines # let us save the first one for name in ['UACC812', 'BT549', 'MCF7', 'BT20']: data = [x[name] for x in nulls] fh = open('%s.json' % name, 'w') import json json.dump(data, fh) fh.close()
-
get_true_prediction()[source]¶ Reads true predcition from the 4 CSV files that contain the true prediction
data is stored as follows in the tru_prediction attribute:
-
get_user_prediction()[source]¶ should be MIDAS files as in https://www.synapse.org/#!Synapse:syn1973835
-
test_synapse_id= 'syn2000886'¶
-
true_synapse_id= 'syn2009136'¶
-
-
class
HPNScoringPrediction_ranking[source]¶ This class is used to compute the ranks of the different participants based on an average rank over the 4 cell lines times phosphos
s = HPNScoringPrediction(filename="file1zip") s.compute_all_rmes() r = HPNScoringPrediction_ranking() # s.aucs is a list where each element is a dictionary of r.add_rme(s1.rmse, "team1") rmse1 = r.get_randomised_rmse(r.rmse[0], sigma=1) rmse2 = r.get_randomised_rmse(r.rmse[0], sigma=2) rmse3 = r.get_randomised_rmse(r.rmse[0], sigma=3) r.add_rmse(rmse1, "team2") r.add_rmse(rmse2, "team3") r.add_rmse(rmse3, "team4") sall.add_rmse(s2.rmse, "team2") sall.get_ranking() {'team1': 1.96875, 'team2': 1.03125}
This class is independent of HPNSCoringPrediction. However, it takes as input the returned values of HPNScoringPrediction.compute_all_rmse()
-
species¶
-
valid_phosphos¶
-
-
class
HPNScoringPredictionInsilico_ranking[source]¶ This class is used to compute the ranks of the different participants based on an average rank over the 4 cell lines times phosphos
s = HPNScoringPredictionInsilico(filename="file1zip") s.compute_all_rmes() r = HPNScoringPrediction_ranking() # s.aucs is a list where each element is a dictionary of r.add_rme(s1.rmse, "team1") rmse1 = r.get_randomised_rmse(r.rmse[0], sigma=1) rmse2 = r.get_randomised_rmse(r.rmse[0], sigma=2) rmse3 = r.get_randomised_rmse(r.rmse[0], sigma=3) r.add_rmse(rmse1, "team2") r.add_rmse(rmse2, "team3") r.add_rmse(rmse3, "team4") sall.add_rmse(s2.rmse, "team2") sall.get_ranking() {'team1': 1.96875, 'team2': 1.03125}
This class is independent of HPNSCoringPrediction. However, it takes as input the returned values of HPNScoringPrediction.compute_all_rmse()
-
class
HPNScoringPredictionInsilico(filename=None, verbose=False, version=2)[source]¶ dimension1 :inhibitor dimenssion2: phosp dimensson3 stimulus dimnesion4 : time
SC2B sub challenge (prediction in silico)
Parameters: - filename – file to score
- version (str) – default to ‘official’ (see note below). Set to anything else to use correct network
Note
This code use the official gold standard used in https://www.synapse.org/#!Synapse:syn1720047/wiki/60532 . Note, however, that a new corrected version is now provided and may be used. Differences with the official version should be small and have no effect on the ranking shown in the synapse page.
-
create_random_data()[source]¶ Here, we don’t want the true prediction that contains only what is requested (AZD8055) but the orignal training data with 2 or 3 inhibitors such as GSK and PD17 so that we can shuffle them.
We want to select for a given cell line and phosphos a data set to fill at a given time. The datum is slected accross the 8 stimuli, inhibitors +DMSO, and time points.
-
test_synapse_id= 'syn2009175'¶
-
true_synapse_id= 'syn2143242'¶
D8C2¶
-
class
D8C2(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D8C2 challenge
from dreamtools import D8C2 s = D8C2() filename = s.download_template() s.score(filename)
Data and templates are downloaded from Synapse. You must have a login.
constructor
-
class
D8C2_sc1(filename, verboseR=True)[source]¶ Scoring class for D8C2 sub challenge 1
from dreamtools impoty D8C2 s = D8C2_sc1(filename) s.run() s.df
see github README for details
-
class
D8C2_sc2(filename, verboseR=True)[source]¶ D8C2 Tox challenge scoring (sub challenge 2)
from dreamtools import D8C2_s2 s = D8C2_sc2(filename) s.run() s.df
see github README for details
DREAM9¶
D9C1¶
Based on https://github.com/Sage-Bionetworks/DREAM9_Broad_Challenge_Scoring/ and instructions and communications from Mehmet Gonen.
Original code in R. Translated to Python by Thomas Cokelaer
-
class
D9C1(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D9C1 challenge
from dreamtools import D9C1 s = D9C1() filename = s.download_template() s.score(filename)
For consistency, all gene essentiality and genomic data files will be given in the same gct file format.
Briefly, this means:
The first and second lines contains the version string and numbers indicating the size of the data table that is contained in the remainder of the file:
#1.2 (# of data rows) (tab) (# of data columns)
The third line contains a list of identifiers for the samples associated with each of the columns in the remainder of the file:
Name (tab) Description (tab) (sample 1 name) (tab) (sample 2 name) (tab) ... (sample N name) And the remainder of the data file contains data for each of the genes. There is one line for each gene and one column for each of the samples. The first two fields in the line contain name and descriptions for the genes (names and descriptions can contain spaces since fields are separated by tabs). The number of lines should agree with the number of data rows specified on line 2.: (gene name) (tab) (gene description) (tab) (col 1 data) (tab) (col 2 data) (tab) ... (col N data)
constructor
D9C2¶
D9C3 scoring function
Based on original source code from Mette Peters found at https://www.synapse.org/#!Synapse:syn4308980
DREAM9.5¶
D9dot5C1¶
D9dot5C1 challenge scoring functions
-
class
D9dot5C1(verbose=True, download=True, **kargs)[source]¶ A class dedicated to D9dot5C1 challenge
from dreamtools import D9dot5C1 s = D9dot5C1() s.download_templates() s.score('templates.txt.gz') # takes about 5 minutes
constructor
-
download_templates()[source]¶ Download a template from synapse into ~/config/dreamtools/dream5/D5C2
Returns: filename and its full path
-